Team:Kyoto/Lab Work
From 2011.igem.org
Lab Work
Week1: Monday 1st - Sunday 7th August
Week2: Monday 8th - Sunday 14th August
Friday
12th
Digestion:transformation of parts listed as below.
- •lactose promoter(ampicilin)
- •double terminator(ampicilin)
- •4-17M:BBa_K325909(chloramphenicol)
- •1-12M:BBa_E0240(ampicilin)
- •2-17F:BBa_120260(kanamycin)
After transformation, we put E.coli in the plate which is containing the each antibiotic.
We failed to conduct transformation of 4-17M,1-12M,and2-17F ,but transformation of lactose promoter and double terminator worked out.
Week3: Monday 15th - Sunday 21th August
Thursday
Digestion:retry of transformation
- We could not work out transformation of 4-17M,1-12M,and2-17F. This was why we tried again.
- However, these experiments were failed.
Digestion:liquid culture of lactose promoter and double terminator
- Picked colonies by using tips ,then put in the LB medium(LB 3ml ampicilin 3μl)
Nutritional Signal(Sugiura,Shimosaka & Okumura)
PCR Amplification of glnL and glnG from gDNA of E.coli
--Primers--
glnL
Left primer: tctagaggagactgctttatggcaac
Right primer: actagtaggaactatcgtcatcgactac
glnG
Left primer: tctagaggtgacgtttatgcaacga
Right primer: actagtacacacaagctgtgaatcactc
annealing temperature was 55 degrees.
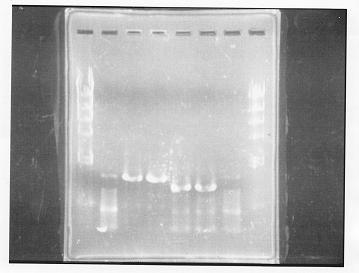
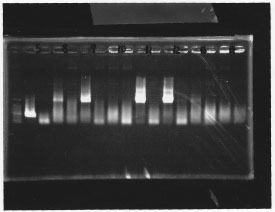
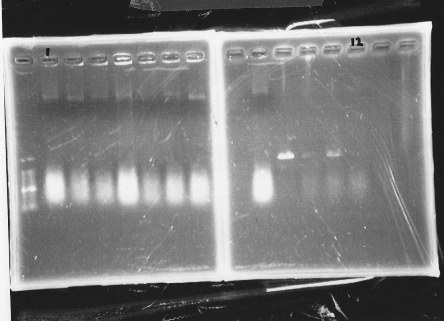
lane 1,2 &7,8: DNA ladder(λDNA digested Hindlll,100bp), lane 3,4: glnG1,glnG2, lane 5,6: glnL1,glnL2
After purification, the concentration of DNA are
glnG1: 127.8ng/ul
glnG2: 118.1ng/ul
glnL1: 137.4ng/ul
glnL2: 124.2ng/ul
Friday
Nutritional Signal(Shimosaka)
Restriction of glnG1 and glnG2
Cut them with Xbal and Spel for 2 hours at 37 degrees.
Then, gel extraction of digested.
Digestion:Mini prep of lactose promoter and double terminator.
- This is results of the experiment.
- •lactose promoter 74.3μl/ml
- •double terminator 6.7μl/ml
- lactose promoter expressed GFD,so we judged this part was not correct one.
Digestion:Transformation of parts listed as below
- 4-17M(luciferage)
- 1-12M(GFP-dT)
- 2-17F(middle copy vector:iGEM 2009kit)
these transformation were failed.
Week4: Monday 22th - Sunday 28th August
Monday
Digestion:Transformation of these parts as below
- 4-17M:BBa_k325909(luciferage)
- 1-12M:BBa_E0249(GFP-dT)
- 2-17F:BBa_I20260(middle copy vector:iGEM 2010kit)
- these transformation were failed.
Tuesday
Digestion:Transformation
- •1-5E(pSB3K3)
Wednesday
Digestion:DNeasy of these parts described as below
- •JCM4616 16.9μg/ml 1.48 260/280
- •JCM5070 7.7μg/ml 1.40 260/280
Digestion:liquid culture
- •double terminator
- •1-5E
Thursday
Digestion:Restriction enzyme digestion
- •double terminator(77.4μg/ml)
- result
- this experiment was failed.
- the reason of this failure was
Capture(Kusaba, Terada, Hara): the experiment about flies' phototaxis ① ♂:UV×2, green×2, ♀:UV×2, green×2
Friday
Capture(Kusaba):the experiment about flies' phototaxis ① ♂:IR×2 ♀:IR×2
Digestion:Miniprep
- •1-5E 7.1μg/ml 1.53 260/280
Digestion:Transformation of a part described as below.
- •1-6G:BBa_R011(pSB1A2)
Digestion:DNeasy of these parts described as below.
- •JCM4616 4.4μg/ml 1.22 260/280 0.28 260/230
- •JCM5070 3.2μg/ml 1.38 260/280 0.25 260/230
Digestion:electrophoresis
- •double terminator(77.4μg/ml)
- •1-5E(7.1μg/ml)
- results
Nutritional Signal(Hashiya):
Transformation of bellow parts.
4-17M:BBa_K325909(lux operon)
1-12M:BBa_E0240
2-17F:BBa_120260(low copy vector)
PCR amplification of
Saturday
Capture(Kusaba):the experiment about flies' phototaxis ① ♂:red×2, blue×2 ♀:red×2, blue×2
Sunday
Week5: Monday 29th August - Sunday 4th September
Tuesday
Digestion: Transformation of 1-5E
Digestion: liquid culture of JCM5070 and JCM4616
Nutritional Signal(Hashiya)
・PCR amplification of glnL and glnG from PCR products,glnL1 and glnG1.
--Primers--
glnL
Left primer: ggaattcgcggccgcttctagaggagactgctttatggcaac
Right primer: ggactagtaggaactatcgtcatcgactac
glnG
Left primer: ggaattcgcggccgcttctagaggtgacgtttatgcaacga
Right primer: ggactagtacacacaagctgtgaatcactc
annealing temperature was 55 degrees.
・PCR amplification of glnL+G and rpoN from gDNA of E.coli.
--Primers--
glnL+G
Left primer: ggaattcgcggccgcttctagaggagactgctttatggcaac
Right primer: ggactagtacacacaagctgtgaatcactc
rpoN
Left primer: ggaattcgcggccgcttctagaggttctgaacatgaagcaa
Right primer: ggactagtatccttatcggttgggtca
annealing temperature was 56 degrees.
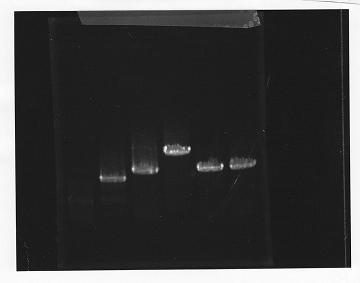
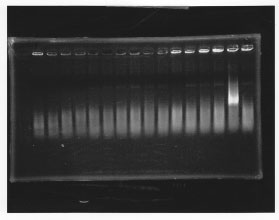
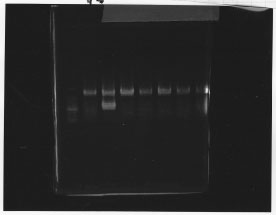
lane1: 100bp DNA ladder, lane2:glnL, lane3:glnG, lane4:glnG+L, lane5:rpoN from gDNA, lane6:rpoN from ASKA clone
After purification, the concentration of DNA are
glnL: 122.3 ng/ul
glnG: 64.7 ng/ul
glnL+G: 106.7 ng/ul
rpoN from gDNA: 111.4 ng/ul
Wednesday
Digestion(nobeyama izumi komatsu):DNeasy of JCM5070 and JCM4616
these results were described as below
JCM5070-1 5.2μg/ml
JCM5070-2 4.9μg/ml
JCM5070-3 4.4.μg/ml
JCM4616-1 10.7μg/ml
JCM4616-2 10.0μg/ml
JCM4616-3 16.2μg/ml
Digestion:liquid culture of 1-6E,JCM5070,and JCM4616
Friday
Digestion: Mini prep of 1-6E-1,1-6E-2,and1-6E-3.
- RESULTS
- 1-6G-1 there were no plasmid.
- 1-6G-2 there were no plasmid.
- 1-6G-3 132.3μg/ml 1.77 260/280 2.16 260/230
Nutritional Signal(Hashiya)
・Restriction of σ54 promoter5,glnL, glnG, glnL+G and rpoN
Cut them with EcoRl and Spel
After purification, the concentration of DNA were
σ54 promoter5: 23.6 ng/ul
glnL: 28.1 ng/ul
glnG: 26.3 ng/ul
glnL+G: 15.3 ng/ul
rpoN: 20.8 ng/ul
・Ligation reaction
Ligated glnL, glnG and rpoN to pSB1K3.
Thursday
Digestion:Mini prep of 1-6E,σ54-5,and σ54-10
- these results were described as below
- 1-6E-1 failure
- 1-6E-2 failure
- 1-6E-3 failure
- σ-54-5 26.4μg/ml 1.70μg/ml 260/280 1.60μg/ml 260/230
- σ-54-10 failure
Digestion:DNeasy of JCM5070 and JCM 4616.
- JCM5070W1 7.1μg/ml
- JCM5070W2 13.6μg/ml
- JCM5070G1 8.1μg/ml
- JCM5070G2 4.8μg/ml
- JCM4616W1 64.9μg/ml
- JCM4616W2 7.5μg/ml
- JCM4616G1 10.1μg/ml
- JCM4616G2 8.1μg/ml
Nutritional Signal(Hashiya)
・Screening PCR of glnL, glnG, glnL+G and rpoN
| glnL | 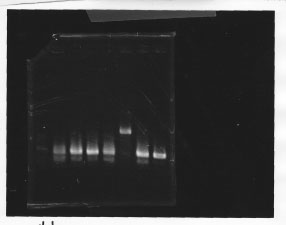 |
| glnG |  |
| glnL+G & rpoN | 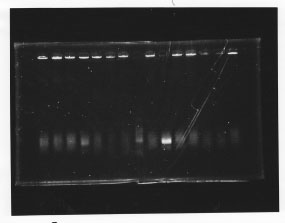 |
We cultured glnL5, glnG4
Saturday
Nutritional Signal(Hashiya)
・Mini prep of glnL5 and glnG4
glnL5: 43.9 ng/ul
glnG4: 38.1 ng/ul
Predation(Hashiya)
・PCR amplification of glmS
--Primers--
left primer:ggaattcgcggccgcttctagagcaggttgaccgacaacgata
right primer:ggactagtacgcagggcatccatttat
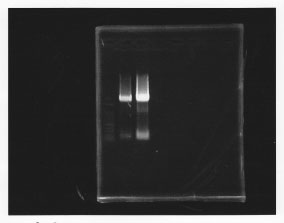
lane1: 100bp DNA ladder, lane2: glmS from gDNA, lane3: glmS from ASKA clone
・TA cloning of glmS
Ligated glmS from gDNA to pTA vector.
Sunday
Nutritional Signal(Hashiya)
・Screening PCR of rpoN
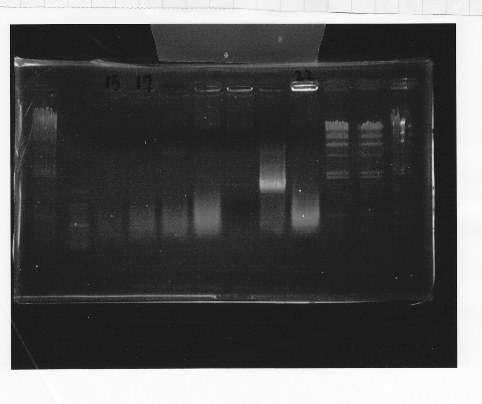
We cultured rpoN-15 and Miniprep, but it did not contain rpoN gene.
・Transformation of bellow parts
1-23L: BBa_B0015 (double terminator)
1-18E: BBa_J23101 (constitutive promoter)
1-18C: BBa_J23100 (constitutive promoter)
Week6: Monday 5th September - Sunday 11th September
Monday
Digestion;retry of liquid culture of 1-6G-1 and 1-6G-2
Tuesday
Digestion: Miniprep of 1-6G which was cultured on September 5
- these results were described as below.
- 1-6G-1 139.1μg/ml 1.63 260/280 2.10 260/230
- 1-6G-2 130.3μg/ml 1.75 260/280 2.19 260/230
Digestion:ethanol precipitation of JCM5070W2(1st September)and JCM4616-3(31th August).
- these results were described as below.
- •JCM5070W2 37.2μg/ml 1.20 260/280 0.65 260/230
- •JCM4616-3 53.0μg/ml 1.13 260/280 0.58 260/230
Capture(Kusaba, Hara):the experiment about flies' phototaxis ②(② is the improved version)♂:green×2 ♀:blue×1
Nutritional Signal(Hashiya)
・Screening PCR of glnL+G(4. sept)
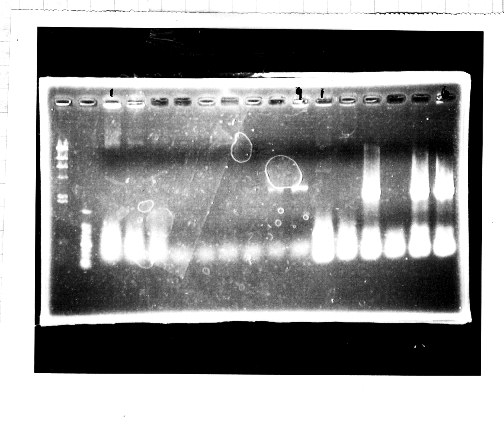
We cultured glnL+G-9.
・Ligation
We ligated rpoN to pSB1C3.
Wednesday
Capture(Hara):the experiment about flies' phototaxis ② ♂:blue×2, green×2, ♀:blue×2, green×2
Nutritional Signal(Hashiya)
・Mini prep of glnL+G-9
The concentration of DNA was 70.4ng/ul.
・Restriction of double terminator and glnL+G-9
Cut double terminator with EcoRl and Xbal.
Cut glnL+G-9 with EcoRl and Spel.
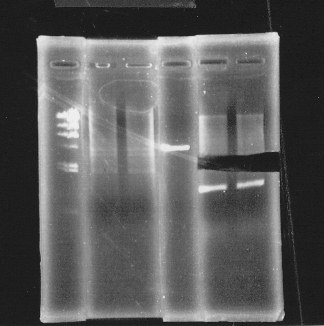
lane1:λ marker, lane2&3:glnL+G cut with E&S, lane4:double terminator cut with E&X, lane5&6:pSB1A3 cut with E&S
Thursday
Capture(Hara):the experiment about flies' phototaxis ② ♂:UV×2 ♀:UV×2
Thursday
Capture(Hara):the experiment about flies' phototaxis ② ♂:UV×2 ♀:UV×2
Nutritional Signal(Hashiya)
・Sequence
We tryed to get sequence of sigma54 promoter-5, glnL-5, glnG-4, rpoN-9 and glnL+G-9. However, they were too thin to get sequence. So, we cultured all of them.
Friday
Nutritional Signal(Hashiya)
・Restriction
Before getting sequence, we cut plasmid to know whether it is correct plasmid.
We cut rpoN-9 with Hindlll, BamHl & Pstl.
We also cut Lux operon with Hindll, EcoRl & Pstl.
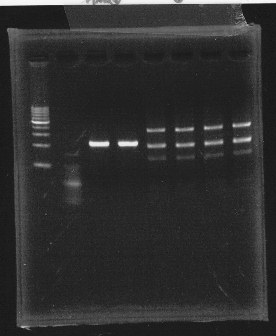
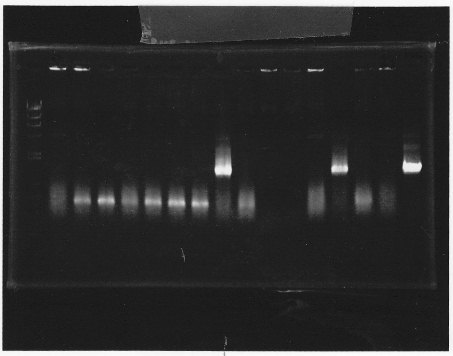
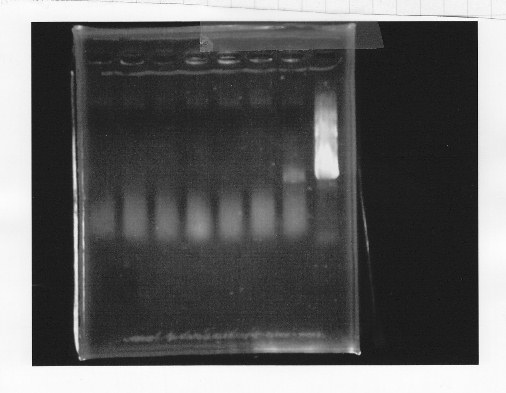
lane1 & 2:1kb DNA ladder and 100bp DNA ladder, lane3,4:digested rpoN-9, lane5,6,7&8:digested lux operon
From this photo, we noticed lux operon were correct but rpoN-9 has some problem.
Saturday
Capture(Kusaba, Hara):the experiment about flies' phototaxis ② ♂:blue×2, green×2, red×2, IR×2, ♀:blue×2, green×2, red×2, IR×2
Nutritional Signal(Hashiya)
・Mini prep
The concentration of DNA were
sigma 54 promoter 135.7 ng/ul
glnL-5 143.2 ng/ul
glnG-4 126.2 ng/ul
glnL+G-9 25.7 ng/ul
rpoN-10 102.0 ng/ul
rpoN-11 95.1 ng/ul
・Restriction
Before getting sequence, we cut plasmid to know whether it is correct plasmid.
We cut rpoN-9, rpoN-10 and rpoN-11 with EcoRl & Pstl.
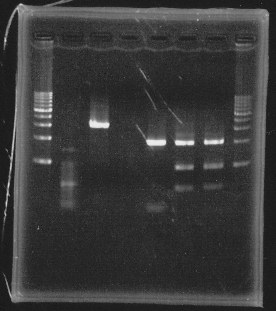
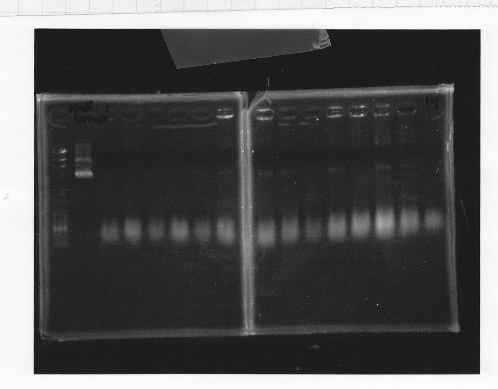
lane1&2:1kb DNA ladder and 100bp DNA ladder, lane5:digested rpoN-9, lane6:digested rpoN-10, lane7:digested rpoN-11
This photo shows rpoN-9 has some problem.
・inverse PCR
To erase restriction site of Pstl in rpoN, we did inverse PCR.
Primer1:gcaagatgccaaatggttgatcaagagtc
Promer2:agattgctgcggataaactgg
Sunday
Capture(Kusaba, Hara):the experiment about flies' phototaxis ② ♂:UV×2
Week7: Monday 12th September - Sunday 18th September
Monday
Capture:transformation of E.coli(Hashiya)
the experiment about flies' phototaxis ②(Kusaba, Hara) ♀:blue×4, UV×2, red×4, IR×4, ♂:blue×2, red×4, IR×2
Nutritional Signal(Hashiya)
・Sequencing
We found glnG-4 is correct. The others, sigma 54 promoter, glnL-5 and glnL+G-9 are not correct.
Tuesday
Capture:E.coli emitted light for the first time. But the amount of light is little.
</div>
Wednesday
Nutritional Signal(Hashiya)
・Mini prep of glnL-9
The concentration of DNA was 136.1 ng/ul
・Sequencing
We found glnL-9 is correct.
Thursday
Nutritional Signal(Hashiya)
・PCR amplification of sigma 54 promoter
We remake sigma 54 promoter.
DNA oligo1:cggaattcgcggccgcttctagagattgca
DNA oligo2:gaatgttgcaccaatatagtgcttcaatggaaacattaagcaccatgttggtgcaatctctagaagcggc
DNA oligo3:gaatgttgcaccaatatagtgcttcaatggaaacattaagcaccatgttggtgcaatctctagaagcggc
DNA oligo4:ggactagtaaaagcgaaatctgtgccaacttttaaattgcccctaaaaggcgttatcatgcgcacc
・Restriction of sigma 54 promoter.
We cut PCR product with EcoRl and Spel.
・Gel extraction of sigma 54 promoter
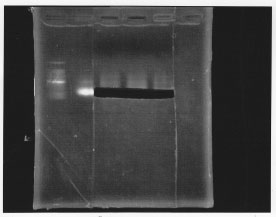
After gel extraction, the concentration of DNA was 18.7 ng/ul.
The concentration of DNA was 136.1 ng/ul
・Screening PCR of rpoN
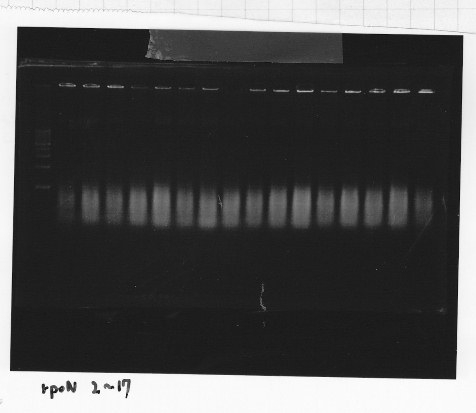
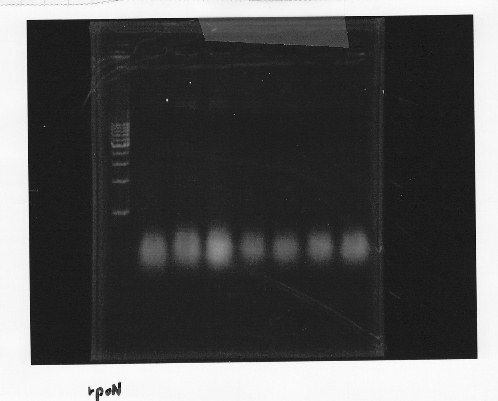
Friday
Digestion(Mori)
PCR amplification of SAM-P20 and ChiA
- We performed colony direct PCRs from a S. albogriseolus colony and a S. avermitilis colony.
Reaction mixture Component Volume(μl) 2x Buffer 25 2mM dNTPs 10 Primer 1 1.5 Primer 2 1.5 Template X KOD FX 1 ddH2O up to 50
PCR condition Predenature 94C 2m Denature 98C 10s 30cycles Annealing 56C 30s Extension 68C 1m30s
- We prepared two kinds of templates:
- Picked a colony, suspended in 50ul of water and incubated 1min at 95 degree. Added 1ul to the reaction mixture.
- Picked a colony and dipped in the reaction mixture.
- Gel electrophoresis indicated that the PCR amplifications were successful for a sample, ChiA, but it was a faint band. We decided to retry direct PCR of SAM-P20 and PCR of ChiA.
Saturday
Digestion(Mori)
Retry of PCR amplification of SAM-P20 and ChiA.
- We amplified SAM-P20 by colony direct PCR and ChiA by PCR using the PCR product that performed yesterday.
- Reaction mixture: The same components and volume as before.
- PCR condition: The same PCR condition as before.
- PCR for SAM-P20
- We prepared two kinds of templates:
- Picked a colony, suspended in 50ul of TE buffer (pH8.0) and incubated 1min at 95 degrees. Added 5ul to the reaction mixture.
- Picked a colony and dipped in the reaction mixture.
- PCR for ChiA
- 1μl of PCR product was added to the reaction mixture as template.
- Gel electrophoresis indicated that the PCR amplifications were successful for all samples. However, an unexpected faint band, about 1000bp, was also observed in the sample of ChiA.
PCR purification of SAM-P20 and gel extraction of ChiA.
- SAM-P20: 43.9 ng/μl
- ChiA: 30.0 ng/μl
Restriction enzyme digestion
- We performed restriction digestions for:
- SAM-P20 with EcoRI and SpeI
- ChiA with EcoRI and SpeI
- Incubated overnight at 37 degrees.
Nutritional Signal(Hashiya)
・Restriction of rpoNm-11
We cut rpoNm-11 with EcoRl and Pstl.
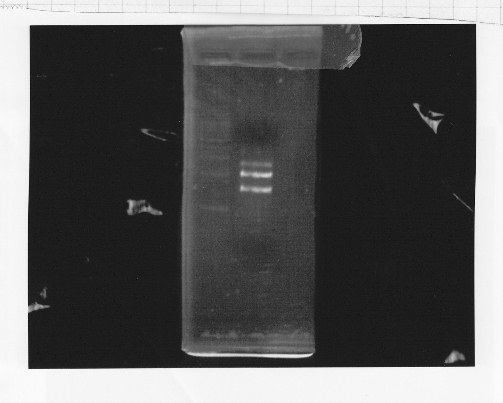
This photo shows the restriction site of rpoN were relieved.
Sunday
Digestion (Mori)
Purification of digested products
- SAM-P20 : 10.2 ng/μl
- ChiA : 22.5 ng/μl
Ligation
Name Vector Insert 1 pSB1C3 SAM-P20 2 pSB1C3 ChiA 3 BBa_B0015 SAM-P20 4 BBa_B0015 ChiA
- Incubated overnight at 16 degrees.
Week8: Monday 19th September - Sunday 25th September
Tuesday
Digestion (Mori)
Transformation
Name Vector Insert Growth 1 pSB1C3 SAM-P20 No 2 pSB1C3 ChiA Yes 3 BBa_B0015 SAM-P20 Yes 4 BBa_B0015 ChiA Yes
Nutritional Signal(Hashiya)
・Restriction of glnL and glnG
We cut glnL and glnG with EcoRl and Spel
・Gel extraction of glnL and glnG
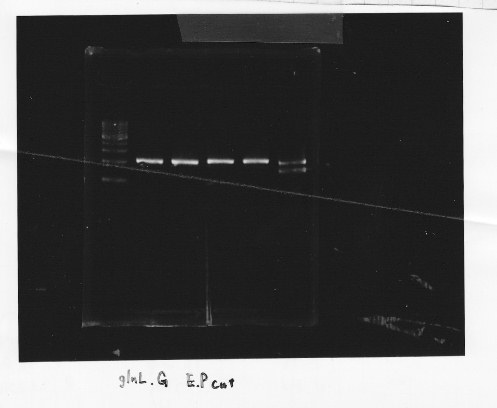
lane1:1kb DNA ladder, lane2&3:glnL, lane4&5:glnG
After gel extraction, the concentration of DNA were
glnL:18.7 ng/ul
glnG:10.0 ng/ul
・Sequencing of rpoNm-11
We found rpoNm-11 is correct.
Wednesday
Digestion (Mori, Nobeyama)
Colony PCR Performed colony PCR to check if ligation and transformation were successful. Gel electrophoresis shown that the ligation and transformation of pSB1C3 with ChiA was successful.
Saturday
Digestion (Hashiya)
・Sequencing analysis
We found sequence of chitinase A1 gene is correct.
Sunday
Capture(Kusaba, Hara):the experiment about flies' phototaxis ② ♂:E.coli×4 ♀:E.coli×4
Digestion (Mori)
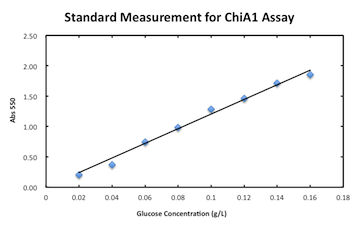
Measurement assay for chitinase A1.
Week9: Monday 26th September - Sunday 2nd October
Monday
Capture(Kusaba, Hara):the experiment about flies' phototaxis ② ♂:E.coli×4, ♀:E.coli×4
 "
"